Model
In this post, we use Monte Carlo simulations to validate the Fama-French five-factor model and the O.L.S properties. We confirm the validity of five factors in the Fama-French model, and uncover some interesting findings of O.L.S properties.
Wikipedia: Fama–French model
- $r$ is the portfolio’s expected rate of return
- $R_f$ is the risk-free return rate
- $K_m$ is the return of the market portfolio
The “five factor” $\beta_2$ to $\beta_6$ are:
- SMB (Small [market capitalization] Minus Big) is the average return on the nine small stock portfolios minus the average return on the nine big stock portfolios;
- HML (High [book-to-market ratio] Minus Low) is the average return on the two value portfolios minus the average return on the two growth portfolios;
- RMW (Robust Minus Weak) is the average return on the two robust operating profitability portfolios minus the average return on the two weak operating profitability portfolios;
- CMA (Conservative Minus Aggressive) is the average return on the two conservative investment portfolios minus the average return on the two aggressive investment portfolios.
They measure the historic excess returns of small caps over big caps and of value stocks over growth stocks.
These factors are calculated with combinations of portfolios composed by ranked stocks (BtM ranking, Cap ranking) and available historical market data. Historical values may be accessed on Kenneth French’s web page.
Here is Description of Fama/French 5 Factors (2x3)
Data Source
Data is downloaded from homepage of Kenneth R. French.
Fama/French 3 Factors [Daily]
This file was created by CMPT_ME_BEME_RETS using the 201801 CRSP database. The 1-month TBill return is from Ibbotson and Associates, Inc.
Portfolios Formed on Book-to-Market [Daily]
This file was created by CMPT_BEME_RETS_DAILY using the 201801 CRSP database. It contains value- and equal-weighted returns for portfolios formed on BE/ME. The portfolios are constructed at the end of June. BE/ME is book equity at the last fiscal year end of the prior calendar year divided by ME at the end of December of the prior year. The annual returns are from January to December. Missing data are indicated by -99.99 or -999. The break points use Compustat firms plus the firms hand-collected from the Moodys Industrial, Utilities, Transportation, and Financial Manuals. The break points include utilities. The portfolios use Compustat firms plus the firms hand-collected from the Moodys Industrial, Utilities, Transportation, and Financial Manuals. The portfolios include utilities.
These portfolios are created by doing the following:
- Rank all stocks that trade on NYSE, Nasdaq and Amex (when it existed) by their book-to-marketratios
- Take the bottom 30% and put them into a portfolio (low 30%)
- Take the next 40% and put them into a portfolio (medium 40%)
- Take the top 30% and put them into a portfolio (top 30%)
- Portfolio weights within each portfolio are value-weights
Load Data
Here is our data:
1 | rm(list=ls()) |
## [1] 2518 26
Extract Fama-French Factors and Fund Returns
rmrf: Km − Rfsmb: SMBhml: HMLrmw: RMWcma: CMArf: Rflo_30: r, the portfolio’s expected rate of return. Here, we choose the portfolio with firms of lowest 30% value- and equal-weighted returns.
Now, we extract Fama-French Factors and Fund Returns from dataset:
1 | mydate <- mydata2$Date |
## [1] "2008-01-02" "2017-12-29"
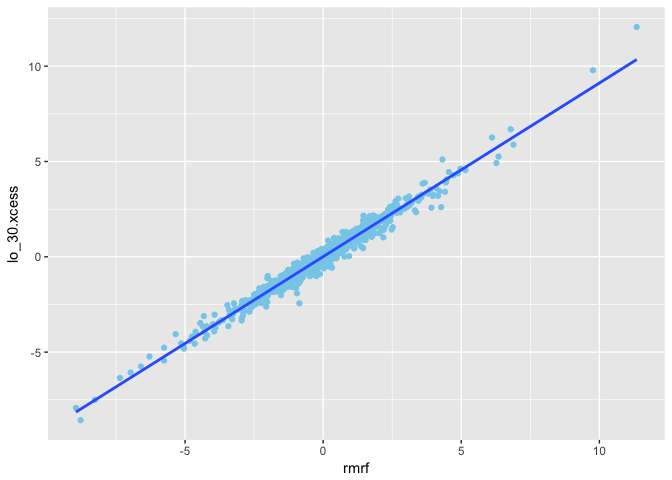
1 | rmrf <- mydata2$Mkt.RF |
The, we calculate Excess Returns for Target fund
1 | lo_30.xcess <- mydata2$Lo.30 - rf |
Run Fama-French Regression
Plot of data:
1 | #========================= |
1 | bwplot(~rmrf, xlab = expression(K[m] - R[f])) |
1 | bwplot(~smb, xlab = "SMB") |
1 | bwplot(~hml, xlab = "HML") |
1 | bwplot(~rmw, xlab = "RMW") |
1 | bwplot(~cma, xlab = "CMA") |
1 | # density plot |
1 | library(car) # red line affected by outlier, green ignoring the outlier (robust) |
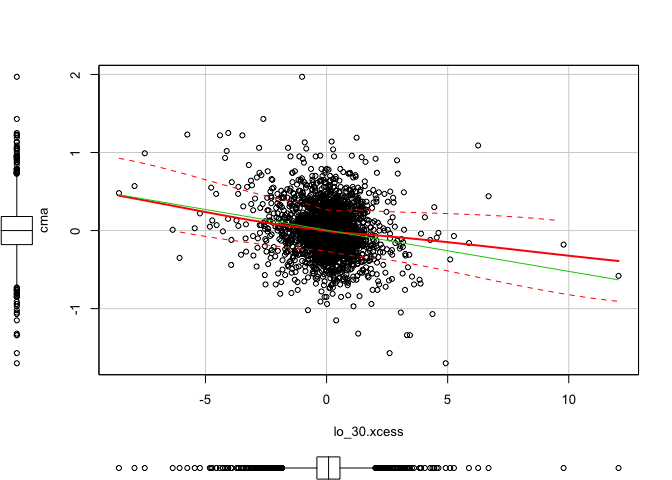
1 | scatterplot(x = lo_30.xcess, y = smb) #(dependent) |
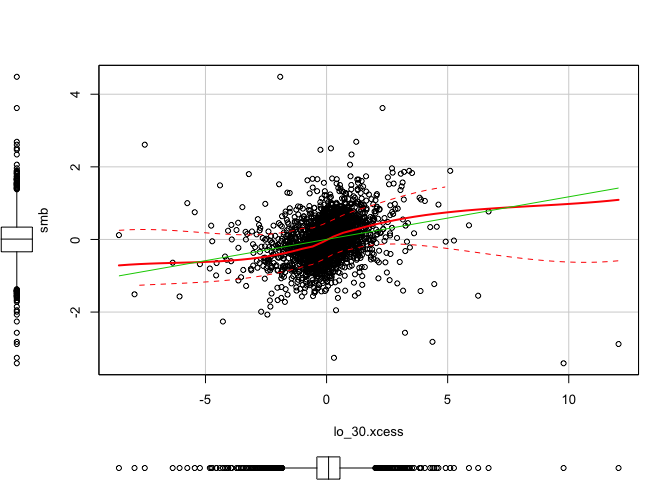
1 | scatterplot(x = lo_30.xcess, y = hml) #(dependent) |

1 | scatterplot(x = lo_30.xcess, y = rmw) #(dependent) |
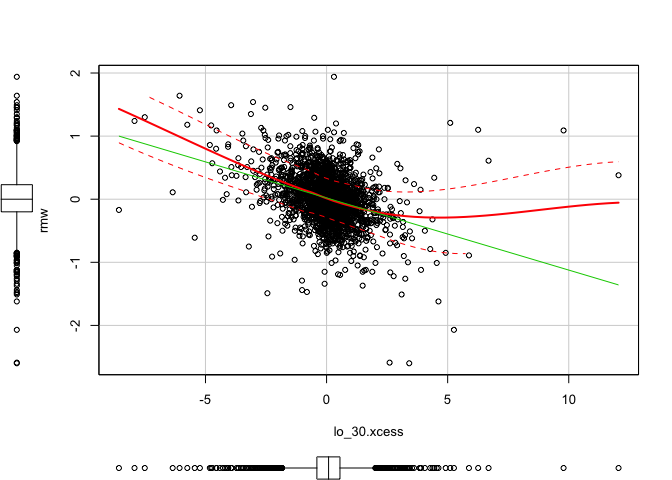
1 | scatterplot(x = lo_30.xcess, y = cma) #(dependent) |
1 | library(ggplot2) |
1 | ggplot(mapping = aes(x = smb, y = lo_30.xcess)) + |
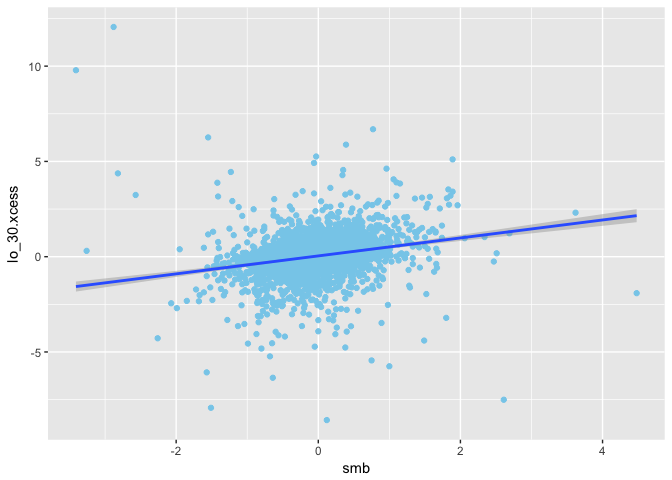
1 | ggplot(mapping = aes(x = hml, y = lo_30.xcess)) + |
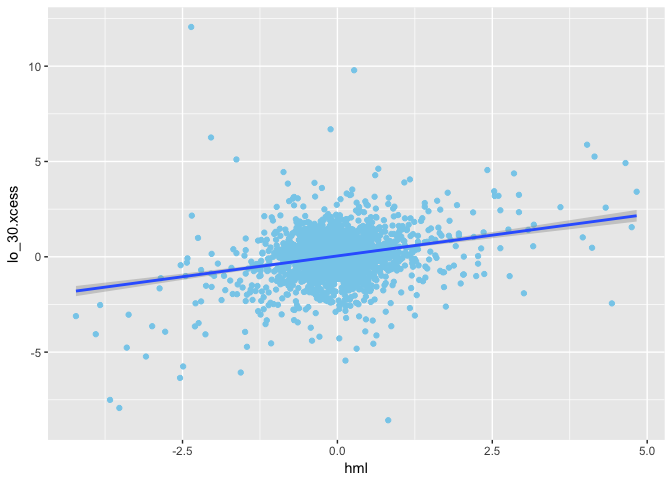
1 | ggplot(mapping = aes(x = rmw, y = lo_30.xcess)) + |
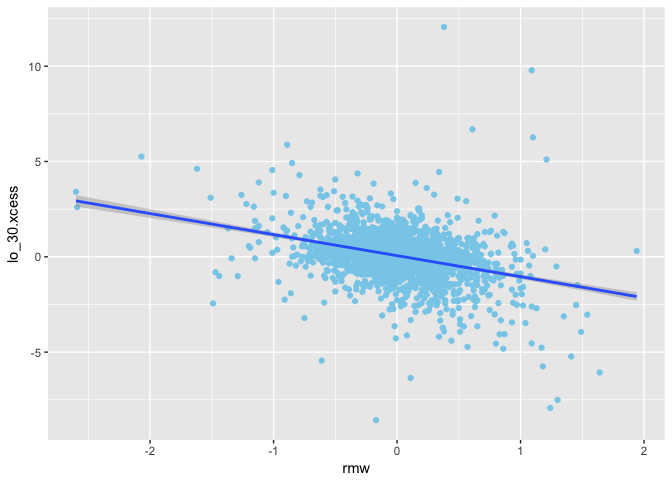
1 | ggplot(mapping = aes(x = cma, y = lo_30.xcess)) + |
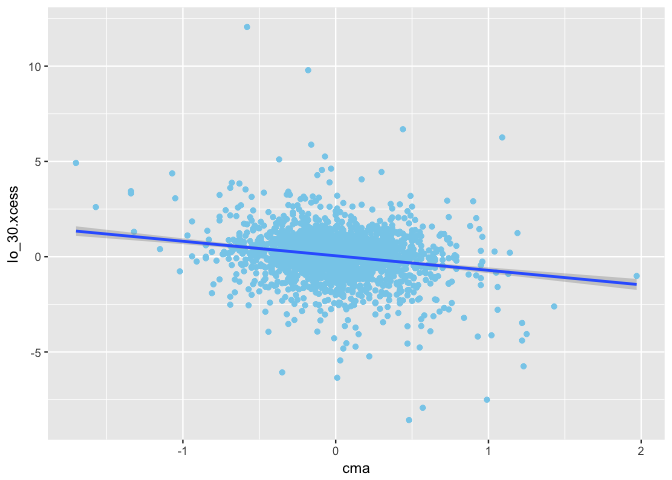
We can run the linear regression to get the β.
1 | lo_30.ffregression <- lm(lo_30.xcess ~ |
##
## Call:
## lm(formula = lo_30.xcess ~ rmrf + smb + hml + rmw + cma)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.59616 -0.05422 -0.00001 0.05556 0.52712
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.004495 0.001949 2.306 0.0212 *
## rmrf 0.979197 0.001806 542.072 < 2e-16 ***
## smb -0.029402 0.003511 -8.374 < 2e-16 ***
## hml -0.258457 0.003373 -76.618 < 2e-16 ***
## rmw 0.055243 0.006215 8.889 < 2e-16 ***
## cma -0.034451 0.006684 -5.154 2.74e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.09759 on 2512 degrees of freedom
## Multiple R-squared: 0.9933, Adjusted R-squared: 0.9933
## F-statistic: 7.486e+04 on 5 and 2512 DF, p-value: < 2.2e-16
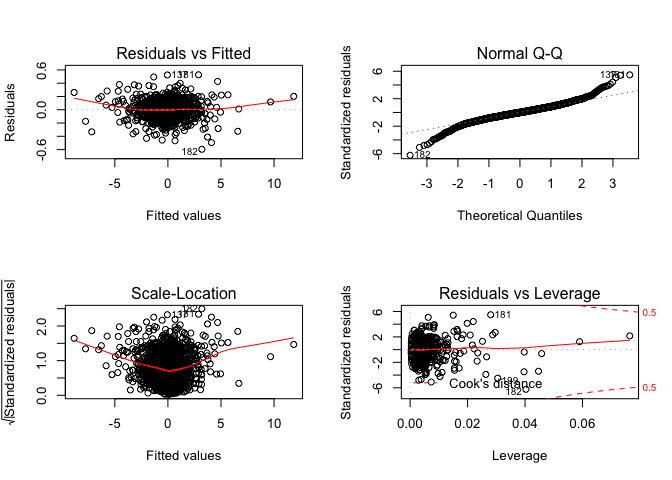
Plot of regression:
1 | par(mfrow=c(2,2)) |
Monte Carlo Simulation
Next, we are running Monte Carlo Simulation for Fama French 5 Factor Model.
1 | #=========================================== |
## [1] TRUE
1 | dim(X) |
## [1] 2518 6
1 | # # obervations |
## [1] TRUE
1 | class(beta) |
## [1] "numeric"
1 | length(beta) |
## [1] 6
1 | # |
## [1] 1.194133
One example of Monte Carlo Simulation
1 | set.seed(12345) |
## [1] "matrix"
1 | is.vector(Y) |
## [1] FALSE
1 | is.matrix(Y) |
## [1] TRUE
1 | length(Y) |
## [1] 2518
1 | ff<-lm(Y~X[,-1]) |
##
## Call:
## lm(formula = Y ~ X[, -1])
##
## Residuals:
## Min 1Q Median 3Q Max
## -3.9939 -0.7897 0.0157 0.8026 3.9580
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -0.002067 0.023685 -0.087 0.930
## X[, -1]rmrf 0.980129 0.021947 44.659 < 2e-16 ***
## X[, -1]smb -0.067441 0.042660 -1.581 0.114
## X[, -1]hml -0.308533 0.040985 -7.528 7.14e-14 ***
## X[, -1]rmw -0.056562 0.075507 -0.749 0.454
## X[, -1]cma -0.023310 0.081204 -0.287 0.774
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.186 on 2512 degrees of freedom
## Multiple R-squared: 0.503, Adjusted R-squared: 0.502
## F-statistic: 508.5 on 5 and 2512 DF, p-value: < 2.2e-16
1 | #============================== |
## 2.5 % 97.5 %
## (Intercept) -0.04851118 0.04437665
## X[, -1]rmrf 0.93709258 1.02316506
## X[, -1]smb -0.15109374 0.01621131
## X[, -1]hml -0.38890087 -0.22816575
## X[, -1]rmw -0.20462411 0.09150049
## X[, -1]cma -0.18254382 0.13592330
1 | class(cfi) |
## [1] "matrix"
1 | dim(cfi) |
## [1] 6 2
1 | # If true beta is in confidence interval of estimated beta |
## [1] TRUE TRUE TRUE TRUE TRUE TRUE
1 | class(flag.beta) |
## [1] "logical"
1 | length(flag.beta) |
## [1] 6
1 | #============================== |
## [1] 1.405962
1 | #============================== |
## (Intercept) X[, -1]rmrf X[, -1]smb X[, -1]hml X[, -1]rmw
## -0.002067263 0.980128819 -0.067441217 -0.308533310 -0.056561812
## X[, -1]cma
## -0.023310260
1 | #============================== |
## (Intercept) X[, -1]rmrf X[, -1]smb X[, -1]hml X[, -1]rmw X[, -1]cma
## FALSE FALSE FALSE FALSE FALSE FALSE
1 | class(flag.type1) |
## [1] "logical"
1 | length(flag.type1) |
## [1] 6
Running Monte Carlo Simulation
Run the Monte Carlo Simulation as follow.
Note:
- this is just a demo with replication number N = 100.
- Actually, we run the Monte Carlo Simulation with replication number N = 105
- We will save the data in a file named
mcff5_2008_2018.RData.
1 | # level of t-test |
Plot
Load the completed Monte Carlo data from file and set the parameters.
1 | rm(list=ls()) |
## [1] "2008-01-02" "2017-12-29"
1 | rmrf <- mydata2$Mkt.RF |
##
## Please cite as:
## Hlavac, Marek (2018). stargazer: Well-Formatted Regression and Summary Statistics Tables.
## R package version 5.2.1. https://CRAN.R-project.org/package=stargazer
1 | stargazer(as.data.frame(X), type = "text", title="Descriptive statistics", digits=4, out="output_star_X.txt") |
##
## Descriptive statistics
## ===============================================
## Statistic N Mean St. Dev. Min Max
## -----------------------------------------------
## V1 2,518 1.0000 0.0000 1 1
## rmrf 2,518 0.0410 1.2911 -8.9500 11.3500
## smb 2,518 0.0080 0.5957 -3.4100 4.4800
## hml 2,518 0.0002 0.7224 -4.2200 4.8300
## rmw 2,518 0.0139 0.3843 -2.6000 1.9400
## cma 2,518 0.0030 0.3146 -1.7000 1.9700
## -----------------------------------------------
1 | # # obervations |
1 | #================================== |
## NULL
1 | #=========================== |
## Adjusted R-squared AIC BIC Mallow's Cp
## Full Model 0.4982286 8046.163 8086.981 NaN
## Model 2 0.1064741 9498.768 9533.756 1963.3724737
## Model 3 0.4981351 8045.635 8080.622 -0.5302520
## Model 4 0.4903992 8084.166 8119.153 38.2578132
## Model 5 0.4981208 8045.706 8080.694 -0.4588785
## Model 6 0.1064741 8045.325 8080.312 -0.8401955
## Model 7 0.4980998 8044.814 8073.970 0.6473720
## Model 8 0.4863740 8100.990 8118.484 61.5142284
1 | #sink() |
## beta1 beta2 beta3 beta4 beta5 beta6
## Full Model 0.04988 0.05079 0.04952 0.05018 0.05194 0.04833
## Model 2 0.29885 NA 0.99999 1.00000 1.00000 1.00000
## Model 3 0.04979 0.05161 NA 0.05018 0.05478 0.04849
## Model 4 0.04823 0.59559 0.08615 NA 0.69012 0.64035
## Model 5 0.04969 0.05283 0.05521 0.05892 NA 0.04890
## Model 6 0.04976 0.05202 0.04929 0.05418 0.05192 NA
## Model 7 0.04977 0.05082 0.05601 0.07178 NA NA
## Model 8 0.04872 0.95811 NA NA NA NA
1 | #sink() |
## beta1 beta2 beta3 beta4
## Full Model 5.097693e-05 4.548356e-06 0.0001277153 0.0000584728
## Model 2 4.978255e-02 NA 0.3205085589 0.6105273388
## Model 3 3.617108e-04 2.554723e-03 NA 0.0025626935
## Model 4 2.947476e-04 4.621952e-02 0.0247329253 NA
## Model 5 8.998804e-04 3.272899e-03 0.0083072828 0.0110698879
## Model 6 1.948356e-04 2.363134e-03 0.0014945671 0.0061072960
## Model 7 7.339447e-04 1.141030e-03 0.0090608459 0.0156089053
## Model 8 3.142667e-03 6.844190e-02 NA NA
## beta5 beta6 Error Variance
## Full Model 0.0001093913 0.0001619033 4.572658e-05
## Model 2 0.6890978267 0.9219254461 1.113663e+00
## Model 3 0.0136155417 0.0041033581 2.201317e-04
## Model 4 0.1751336438 0.1788627543 2.221245e-02
## Model 5 NA 0.0060159119 2.605182e-04
## Model 6 0.0032572051 NA 4.449708e-05
## Model 7 NA NA 3.202380e-04
## Model 8 NA NA 3.365506e-02
1 | #sink() |
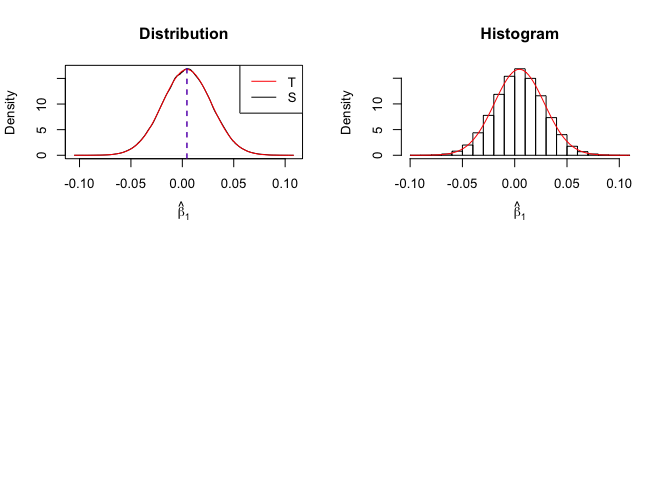
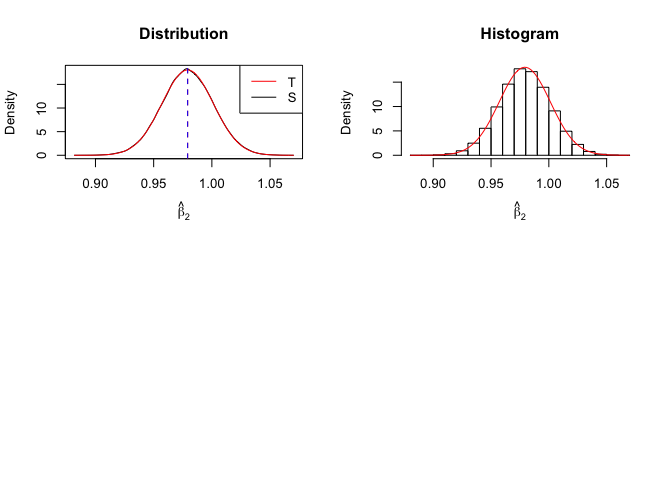
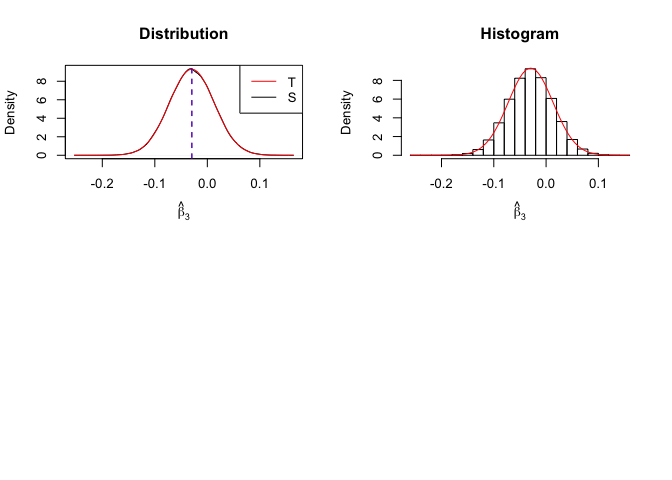
Plot of density of estimator
Number of replication N = 105
1 | # In Distribution pic, blue line is true beta, red line is betahat |
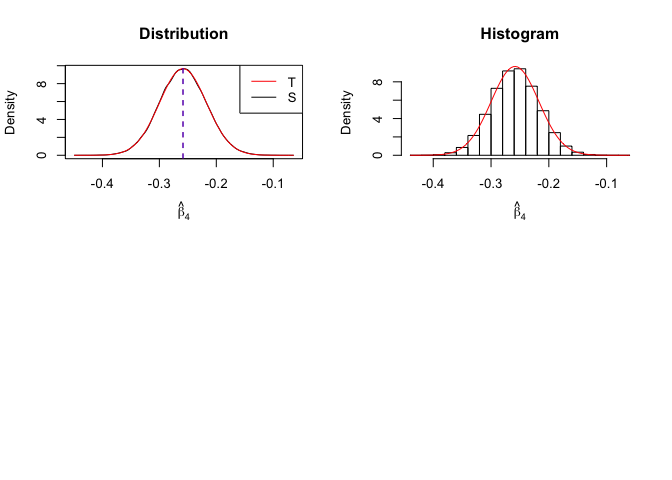
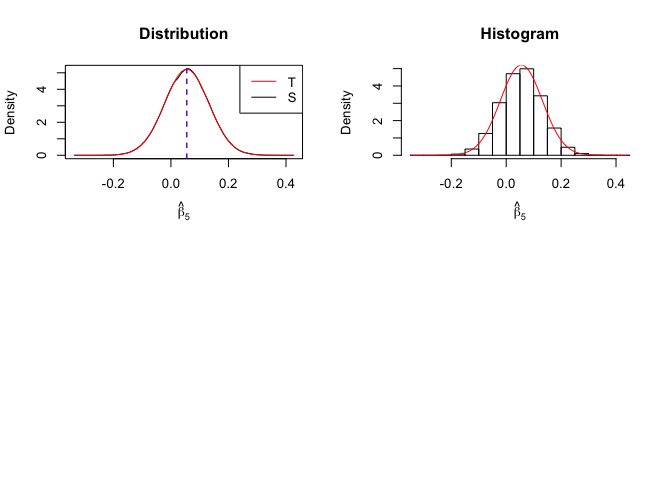
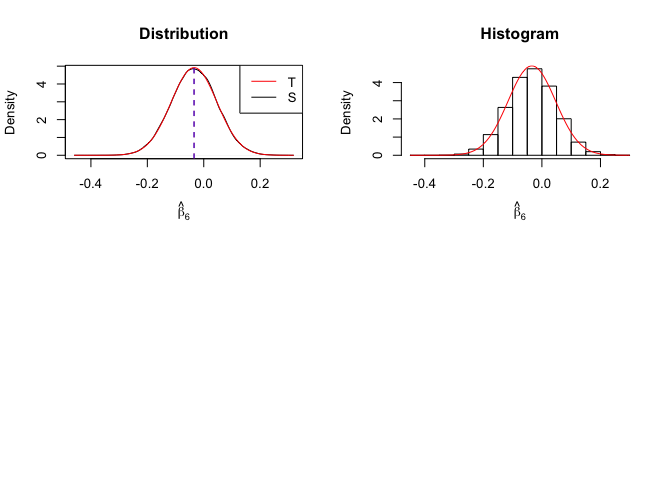
Finding (i)
- The O.L.S. estimators of the unknown coefficients and error variance are unbiased
Plot of betahat
1 | foo = expression(hat(beta)[1], hat(beta)[2],hat(beta)[3], |
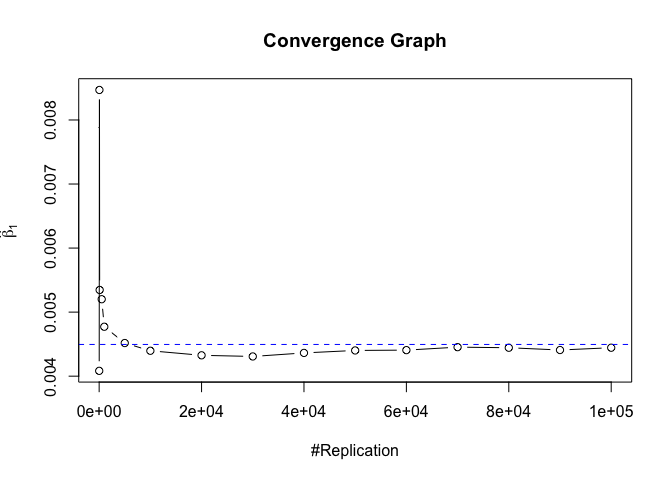
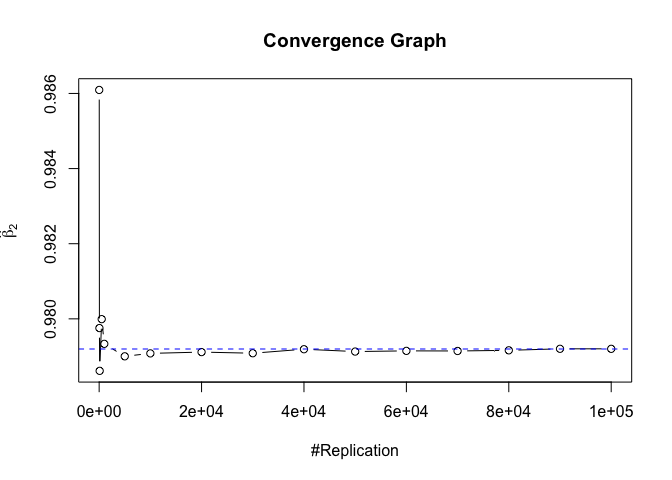
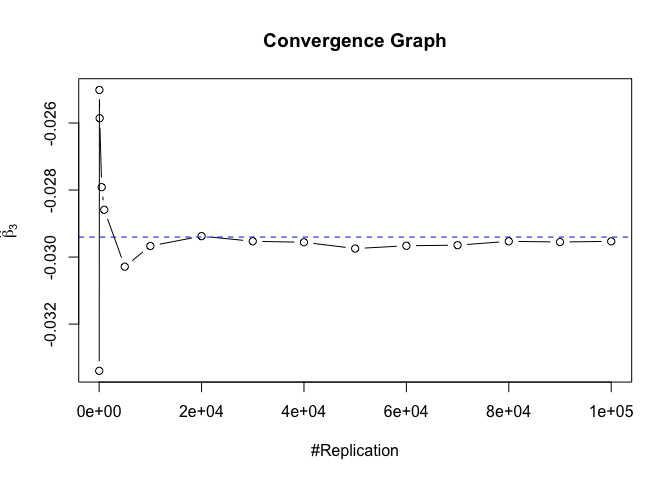
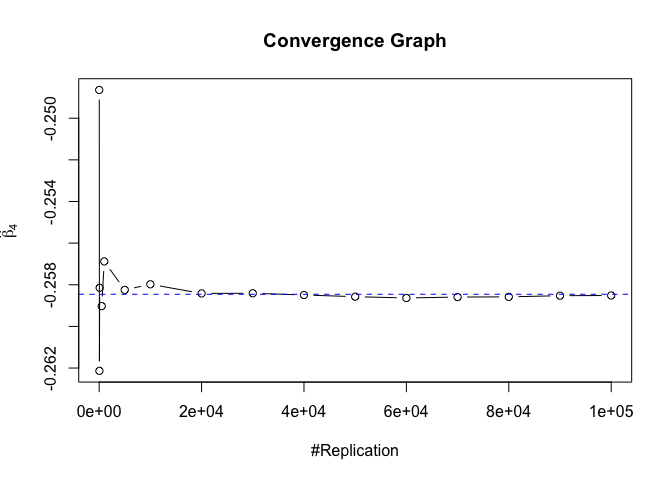
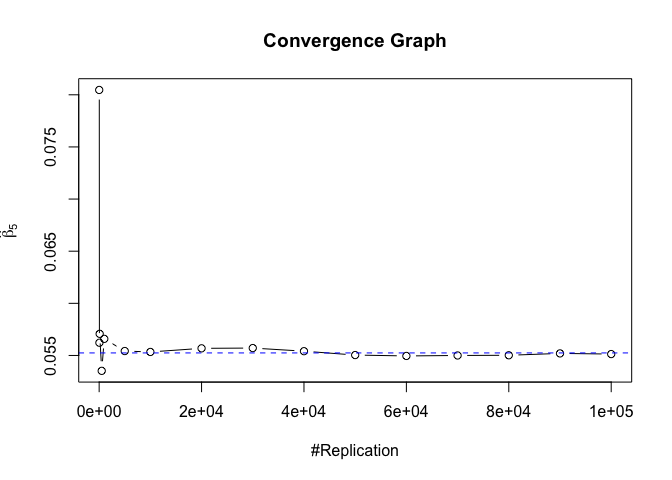
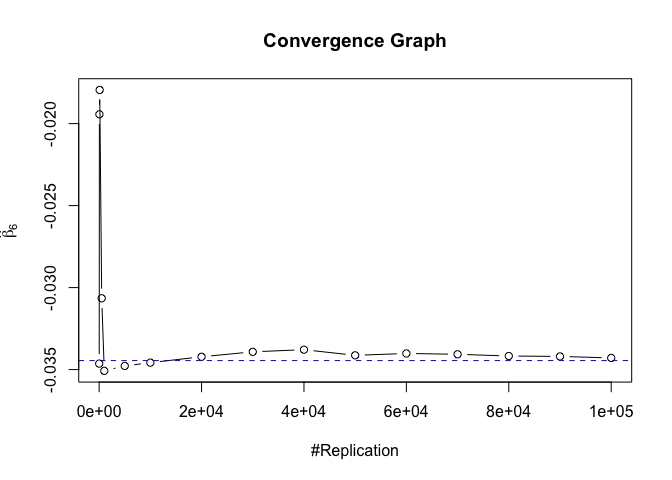
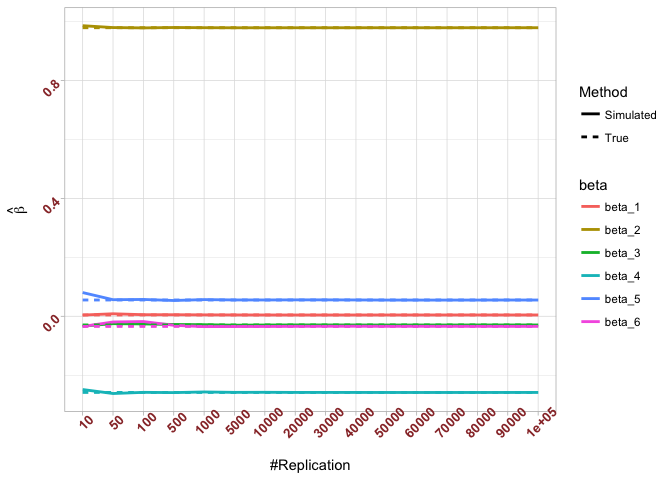
Plot in one graph:
1 | # In one graph |
The difference between true value and the last similated value
1 | abs(beta-mat.betahat[length(vecN),]) |
## (Intercept) rmrf smb hml rmw
## 5.097693e-05 4.548356e-06 1.277153e-04 5.847280e-05 1.093913e-04
## cma
## 1.619033e-04
Plot of variance of betahat
1 | # beta - betahat ~ N(0, sigma^2 (X^T X)^-1) |
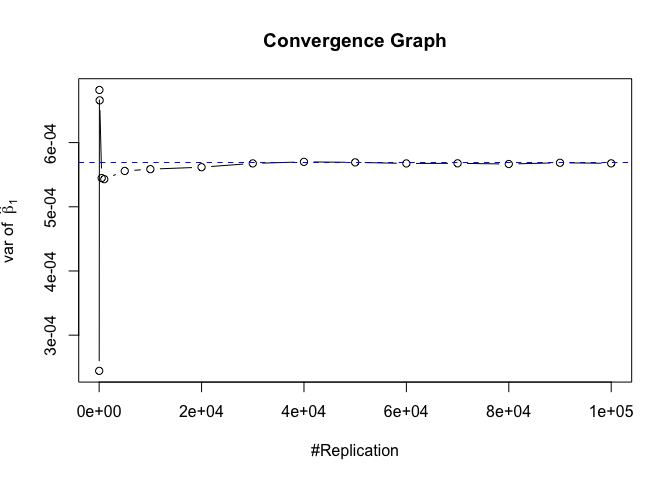
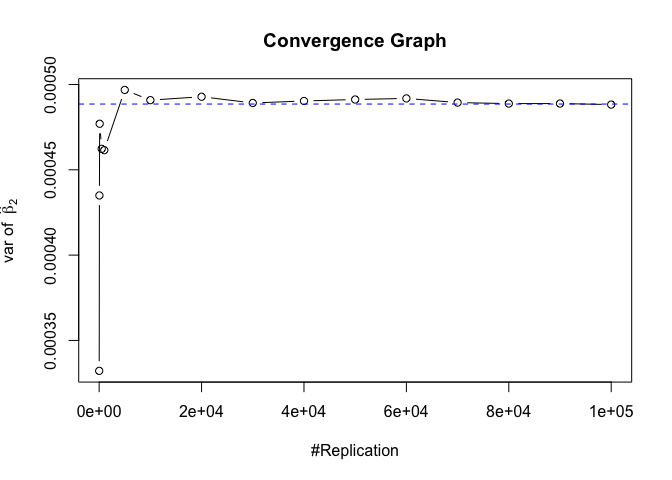
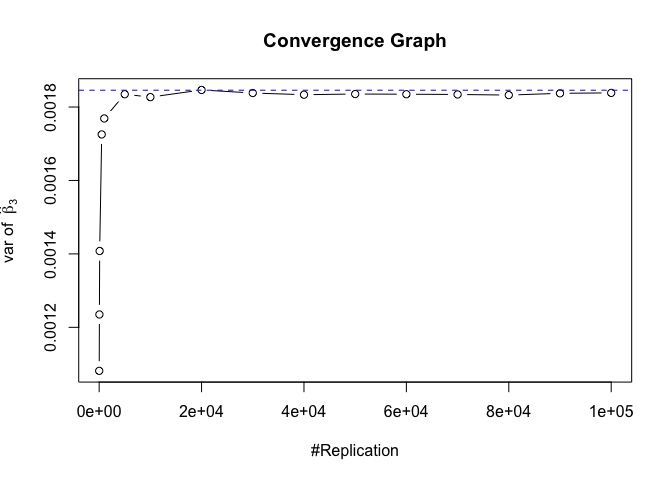
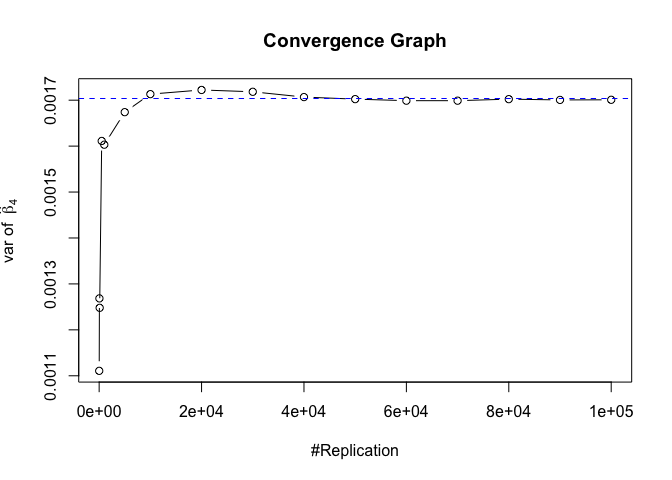
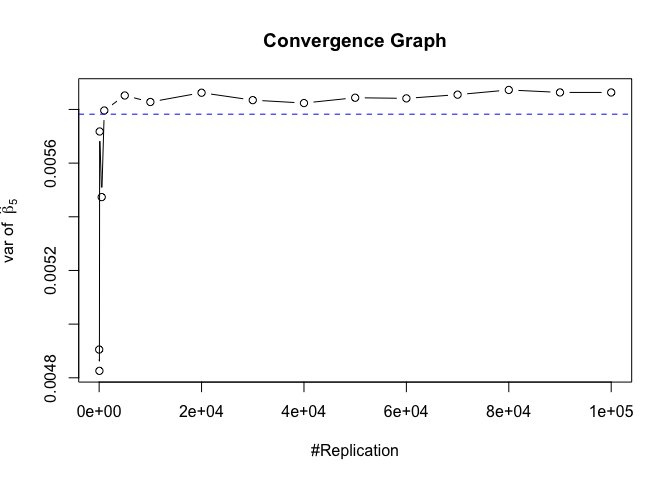
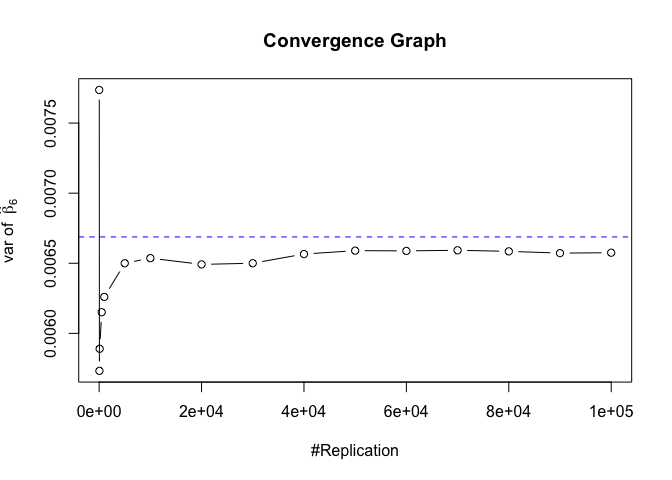
The difference between true value and the last similated value
1 | abs(diag(var.beta)-mat.vebh[length(vecN),]) |
## rmrf smb hml rmw
## 1.275990e-06 3.146077e-07 7.275822e-06 2.791952e-06 8.092901e-05
## cma
## 1.124775e-04
Plot of error variance
1 | # Error Variance |
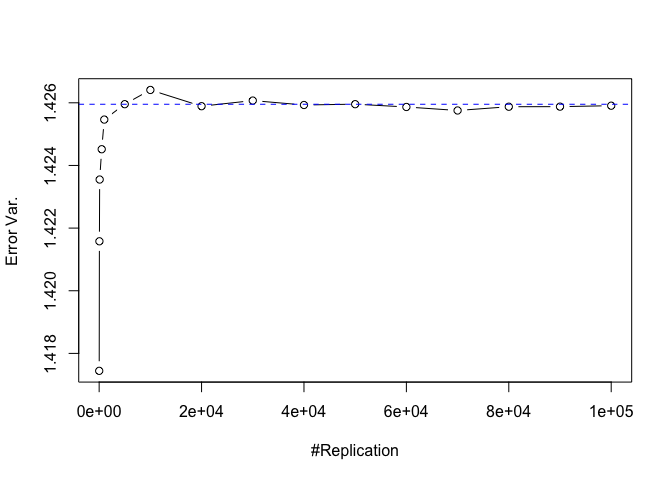
1 | # dev.off() |
The difference between true value and the last similated value:
1 | abs(sig2e-vec.err.varh[length(vecN)]) |
## [1] 4.572658e-05
Finding (ii)
- The “correct” meaning of a 100(1‐α)% confidence interval of an unknown coefficient
1 | for (i in 1:length(beta)){ |
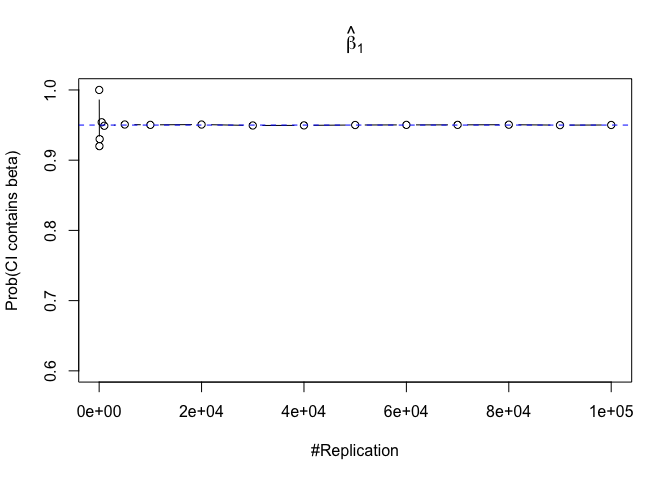
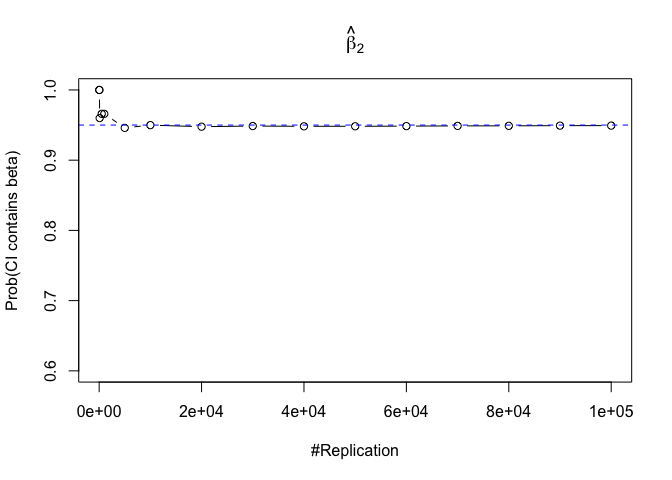
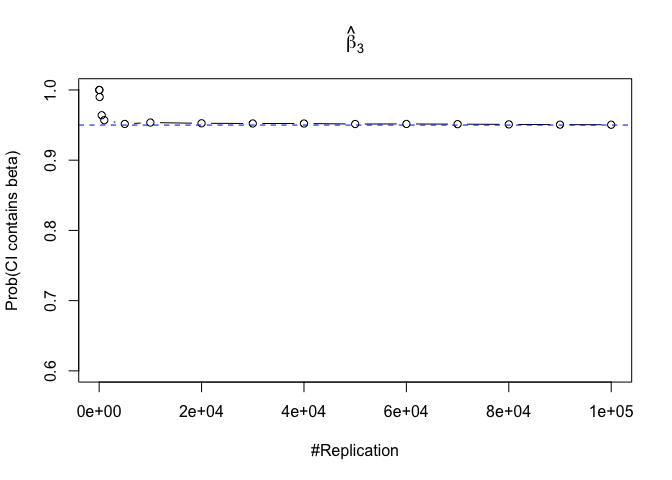
Plot in one graph:
1 | # in one graph |
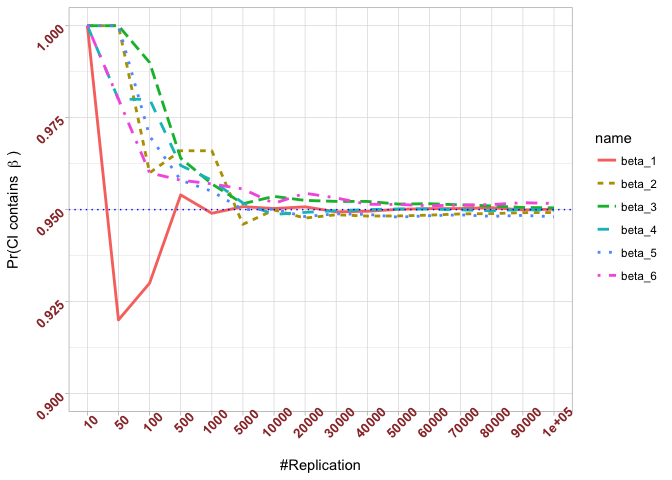
The difference between true value and the last similated value
1 | abs(0.95-prob.flag.beta[length(vecN),]) |
## beta_1 beta_2 beta_3 beta_4 beta_5 beta_6
## 0.00012 0.00079 0.00048 0.00018 0.00194 0.00167
Finding (iii)
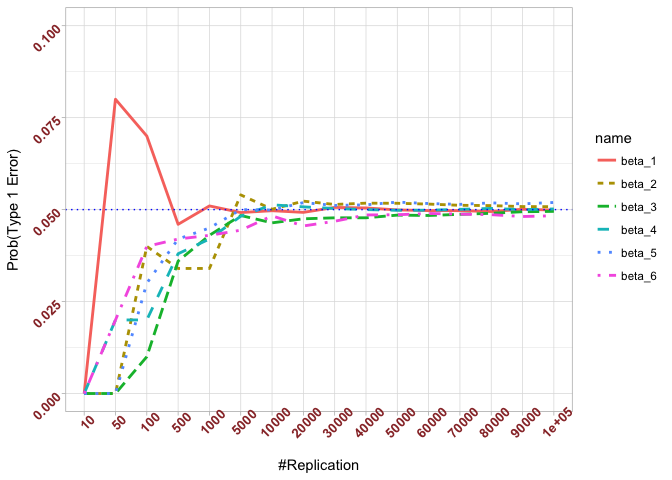
- The significance level of the t test for testing a linear hypothesis concerning one or more coefficients is the probability of committing a Type I error
1 | for (i in 1:length(beta)){ |
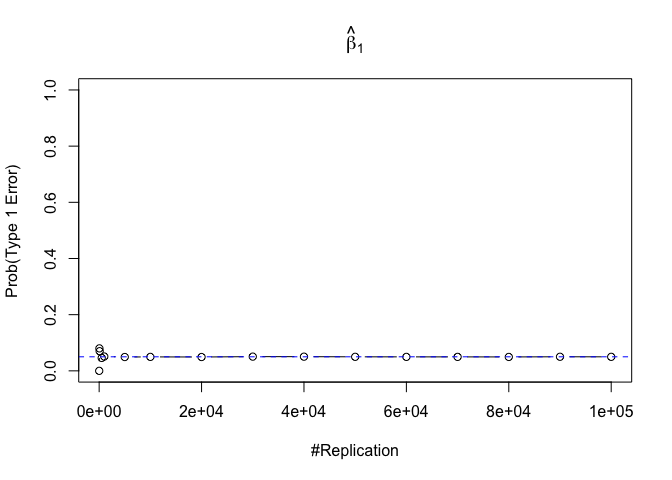
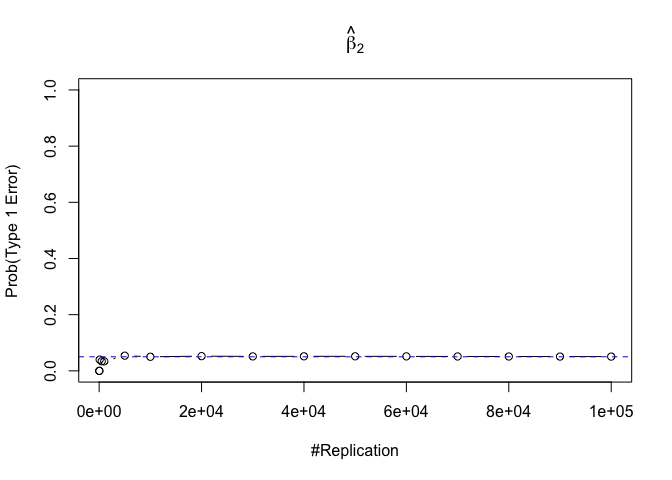
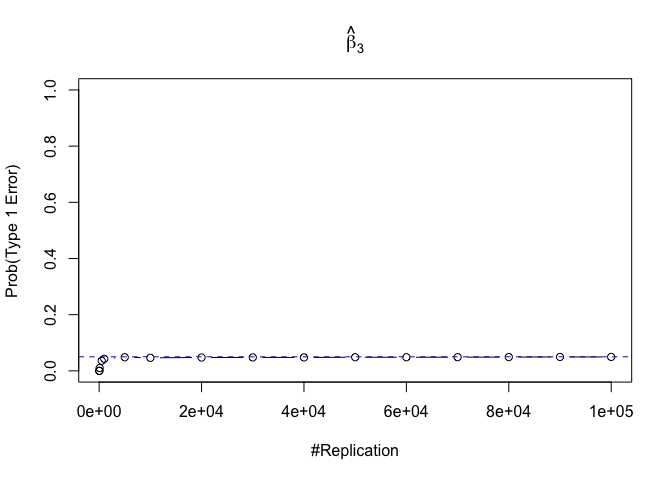

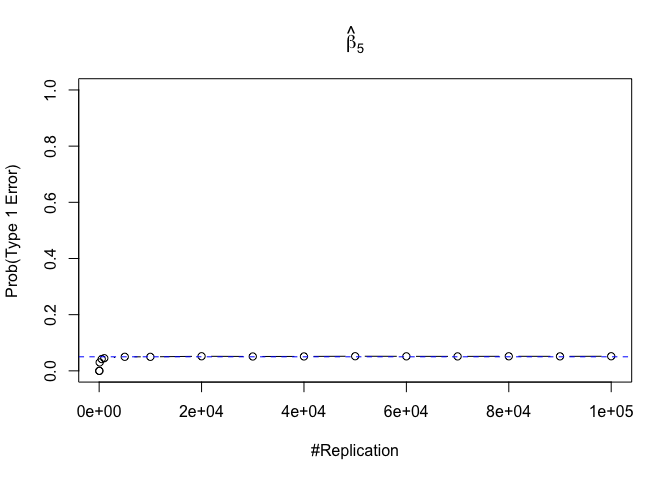
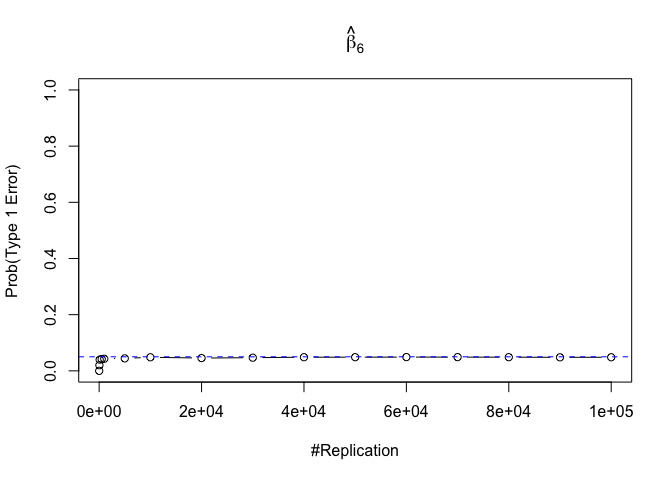
Plot in one graph:
1 | # in one graph |
The difference between true value and the last similated value
1 | abs(0.05-prob.flag.type1[length(vecN),]) |
## beta_1 beta_2 beta_3 beta_4 beta_5 beta_6
## 0.00012 0.00079 0.00048 0.00018 0.00194 0.00167
Finding (iv)
- The t test is unbiased
1 | for (i in 1:length(beta)){ |
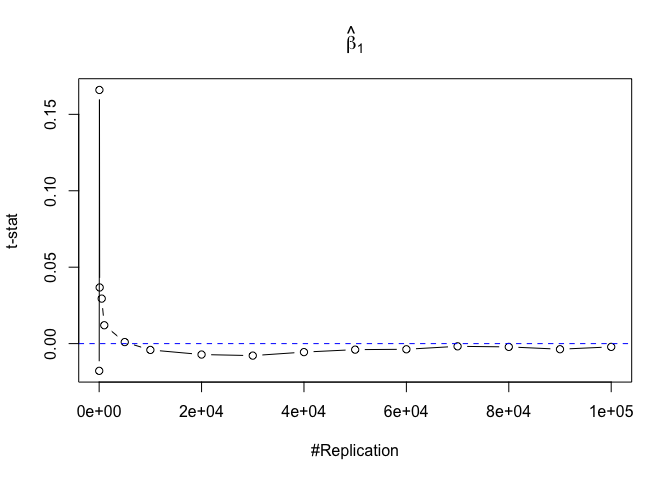
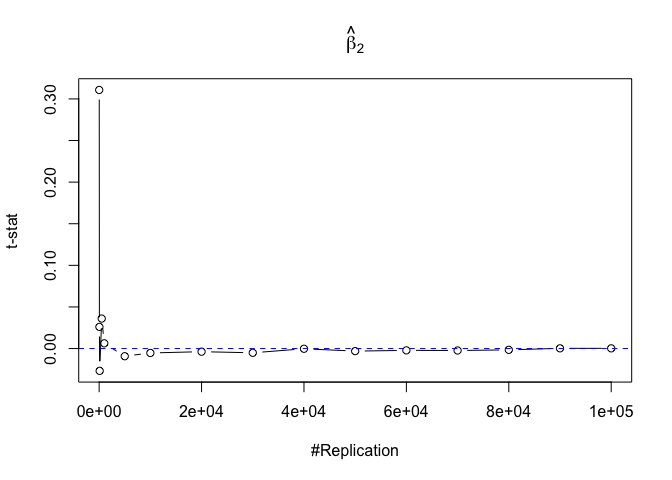
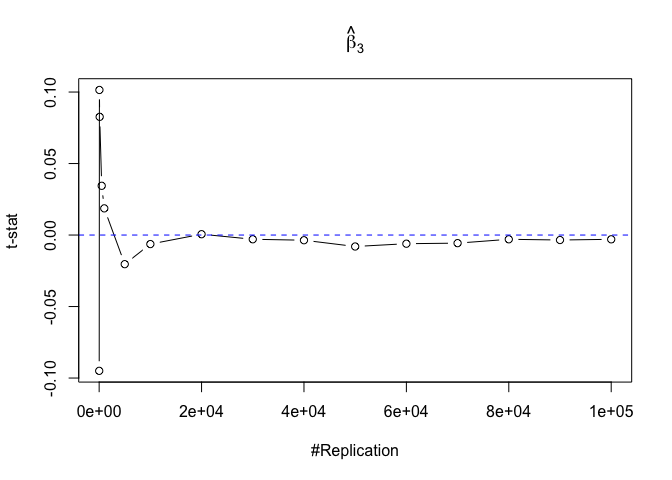
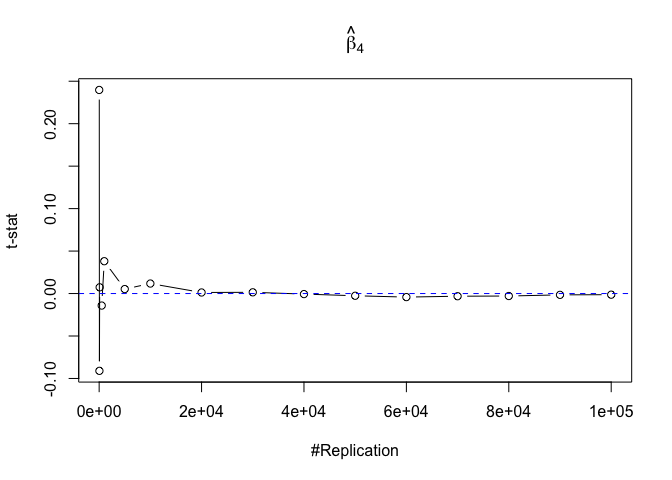
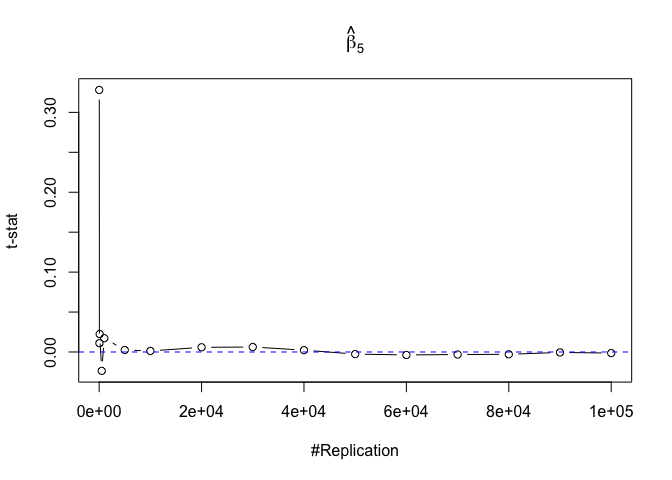
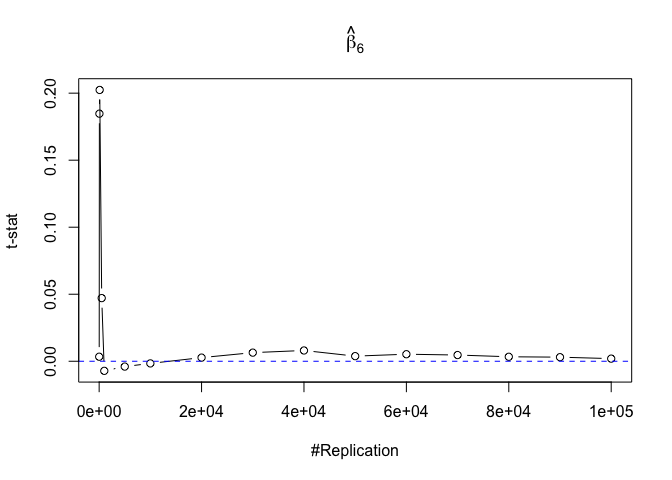
Plot in one graph:
1 | # in one graph |
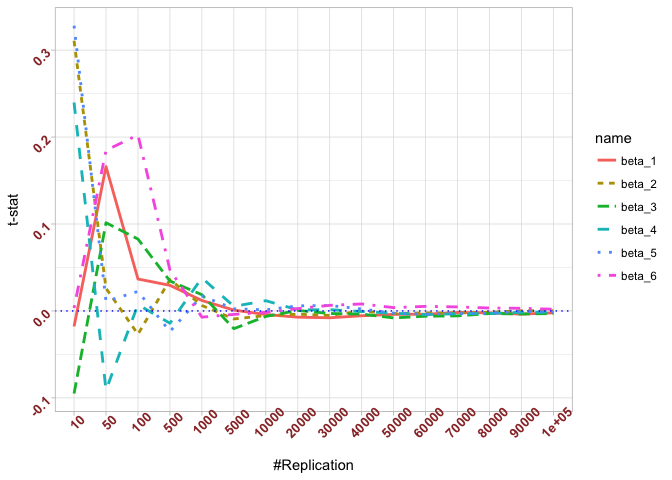
The difference between true value and the last similated value
1 | abs(0-mat.t.stathat[length(vecN),]) |
## beta_1 beta_2 beta_3 beta_4 beta_5
## 0.0021438691 0.0002507301 0.0029878315 0.0013417811 0.0013706685
## beta_6
## 0.0019628747
Finding (v)
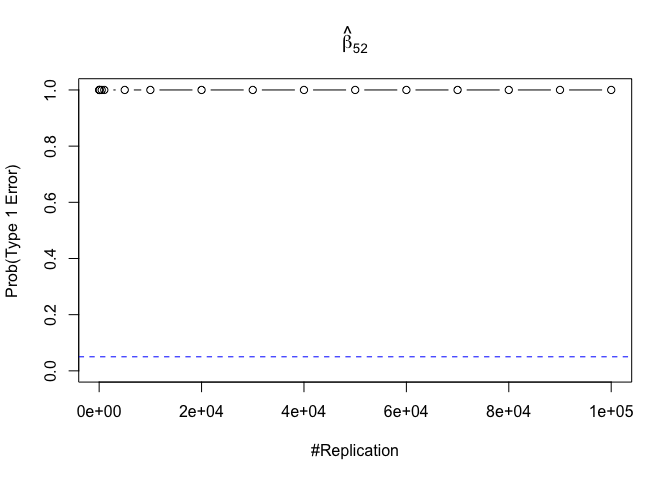
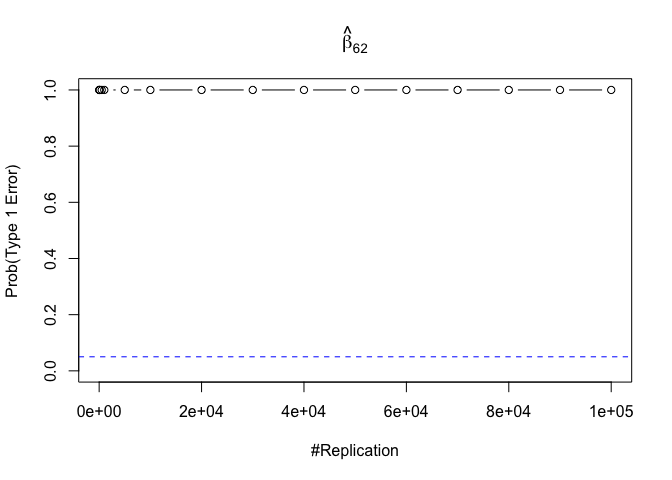
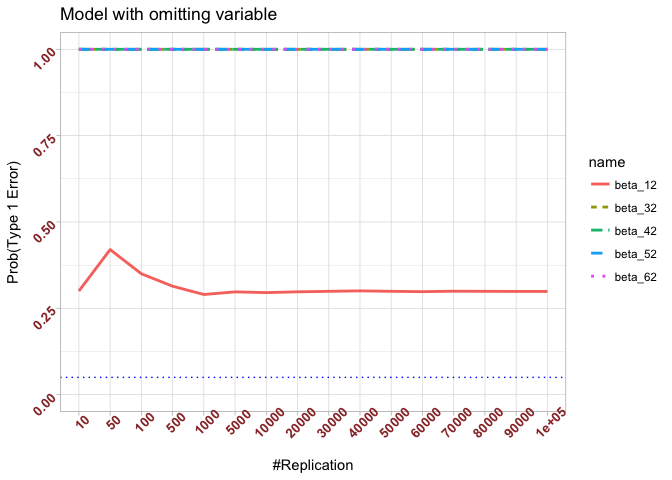
- The result in Part iii) no longer holds if some relevant explanatory variables have been omitted from the model
We omitt the variable rmrf (Km − Rf).
1 | foo3 = expression(hat(beta)[12], hat(beta)[32],hat(beta)[42],hat(beta)[52],hat(beta)[62]) |
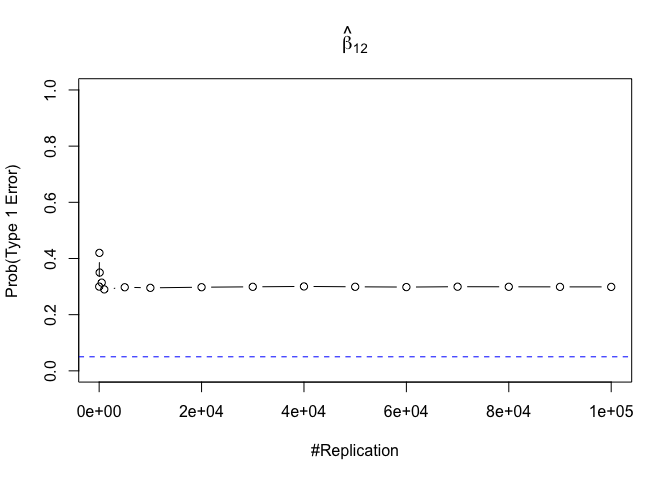
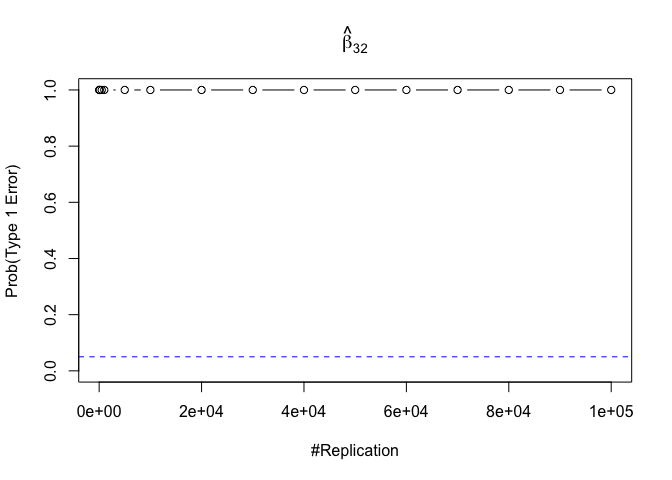
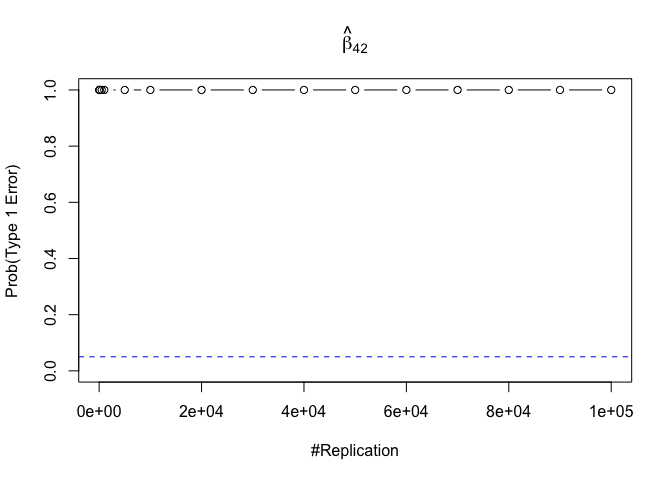
1 | # in one graph |
The difference between true value and the last similated value
1 | abs(0.05-prob.flag.type1.2[length(vecN),]) |
## beta_12 beta_32 beta_42 beta_52 beta_62
## 0.24885 0.94999 0.95000 0.95000 0.95000
Finding (vi)
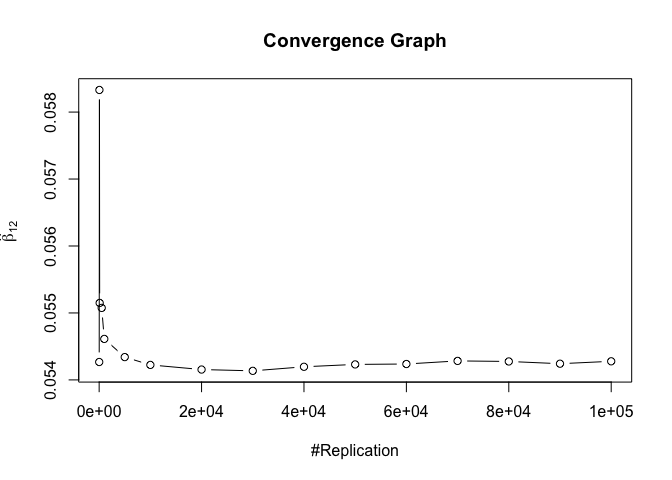
- The estimator of say, the coefficient of X2, is no longer unbiased if the decision of whether to include X1 in the model is dependent on the outcome of a t test. Based on your findings, discuss the wider implications of “model selection” for statistical modeling and the lessons to be learnt for practitioners.
Plot of betahat of omitting model
1 | temp <- 1:length(beta) |
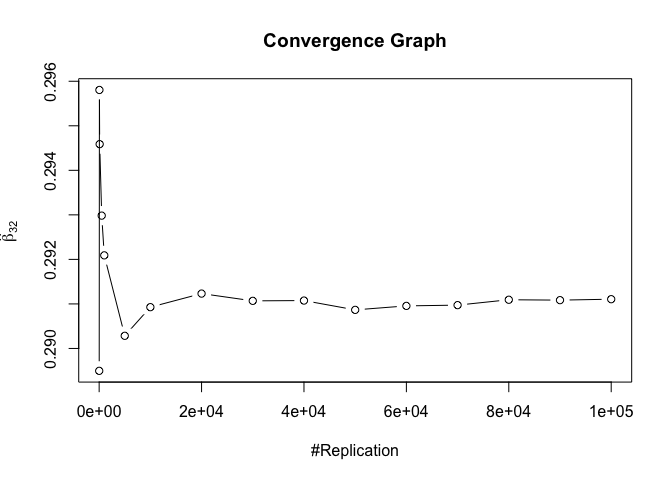
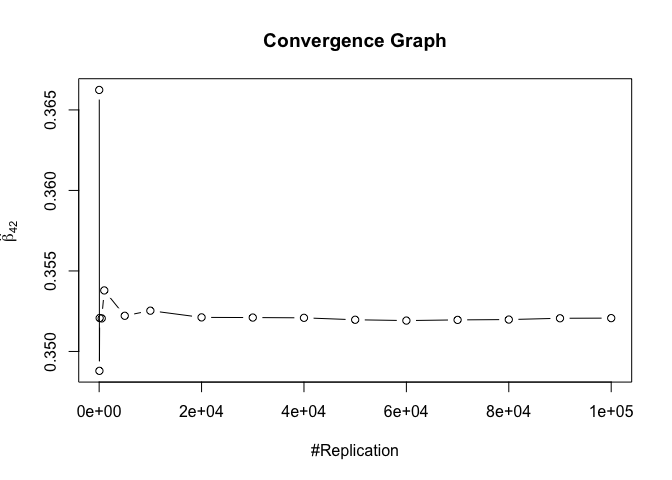
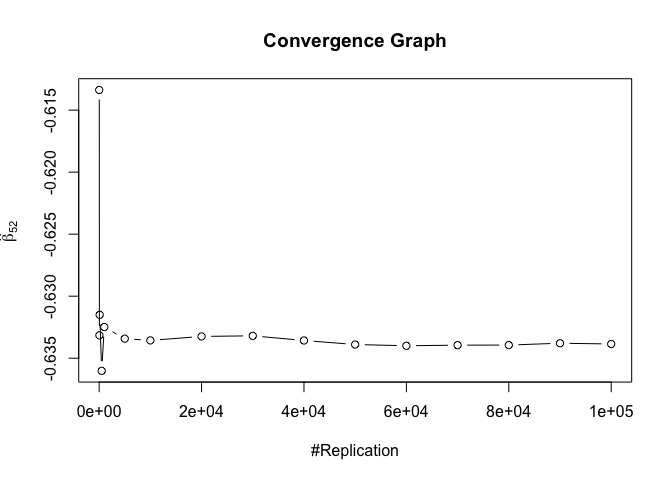
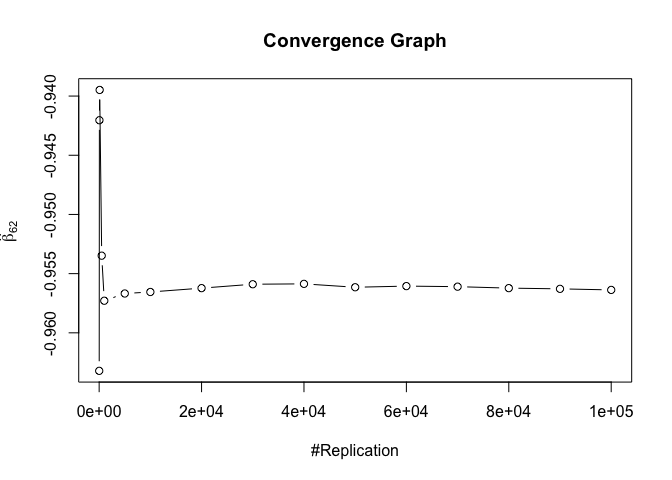
1 | # One graph |
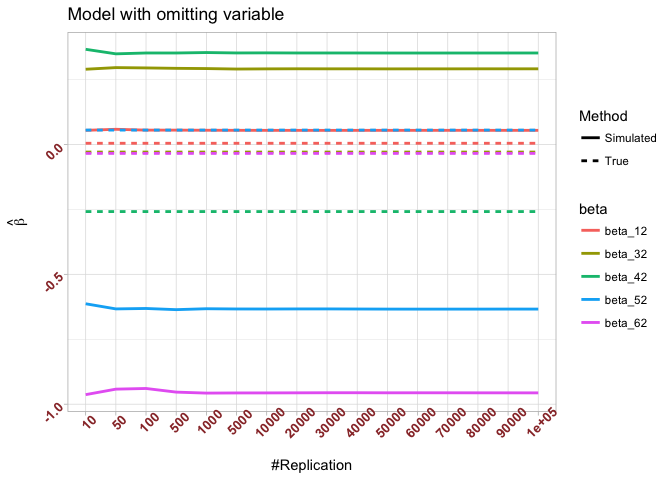
The difference between true value and the last similated value
1 | abs(beta[nbv]-mat.betahat2[length(vecN),]) |
## (Intercept) smb hml rmw cma
## 0.04978255 0.32050856 0.61052734 0.68909783 0.92192545
Plot of Error Variance of omitting model
Unbised estimater of sigma^2
1 | # mypath <- file.path(getwd(),"Figure","vi_var_err.jpeg") |

1 | # dev.off() |
The difference between true value and the last similated value
1 | abs(sig2e-vec.err.varh2[length(vecN)]) |
## [1] 1.113663
Reference
Monte Carlo Method
- Monte Carlo Methods Lecture Notes by Adam M. Johansen and Ludger Evers, November 15, 2007
- Machine Learning: The elegant way to extract information from data: Lecture notes on Regression: Markov Chain Monte Carlo (MCMC)
- Monte Carlo Simulation Basics, I: Historical Notes
- Monte Carlo Simulation Basics, II: Estimator Properties
- Monte Carlo Simulation Basics, III: Regression Model Estimators
Fama French Model
Intro
- Fama-French three-factor model
- How to Download Portfolios and Factors from Fama and French Directly into R
- Estimate the Fama and French Three Factor Model
- Screencast: Fama-French Regression Tutorial Using R
- ken.french
- Could someone teach me how to construct the portfolios by compute (like using R, Excel or Eviews)
Tutorial
- How to use the Fama French Model
- Empirical tests of Fama-French three-factor model and Principle Component Analysis on the Chinese stock market
- Fama-French Multi-Factor Models
- alexpetralia/fama_french
- Description of Fama/French Factors
- Detail for 6 Portfolios Formed on Size and Book-to-Market
- shabbychef/MarkowitzR
- FRE 7241 Algorithmic Portfolio Management
